
The members of the class Microsporea in the phylum Microspora are commonly called microsporidia. Microsporidia are obligately intracellular organisms that are able to infect numerous vertebrate and invertebrate species. They possess unicellular spores, containing a uninucleate or binucleate sporoplasm, and an extension apparatus which always has a polar filament and a polar cap. They do not have mitochondria, and their ribosomal RNAs have prokaryotic properties.
Nosema bombycis Nageli is one of the most investigated microsporidia in Japan, since it is pathogenic to silkworm (Bombyx mori Linnaeus). Nosema means disease, and bombycis originates from the scientific name of silkworm. The disease caused by the microsporidia is called pebrine from pebre, which is the provincial name for pepper in French. In pebrine of the silkworm, the dark mottled areas or darkbrown spots on the integument are a sign of this disease. Definitive species identification and classification can be accomplished using size, shape, ultrastructural examination of spore by light and electron microscopy, life cycle in insect cell lines, spore surface antigen detection by immunological methods (enzyme linked immunosorbent assay, antibody - sensitized latex agglutination test, fluorescent antibody test), host range and target tissue by inoculation tests, spore germination test using chemicals and so on.
Since the first report concerning the extraction of DNA from microsporidian spores, there has been an increasing number of molecular studies of this group of protoctist parasites. A majority of these have focused on determining the nucleotide sequences of ribosomal RNA and their ITS region. These results are being used to investigate theories of early eukaryote evolution and, on a more practical level, to assist in species identifications. Until now, these molecular studies focused on the interspecies diversity, but there were few studies about the intraspecies diversity.
Fig. 1
Immunoblotting analysis of spores protein with antisera to whole spore of Nosema bombycis NIS 001. Proteins extracted from spores of Nosema bombycis NIS 001 (lane 1), NIS 408 (lane 2), NIS 520 (lane 3), NIS 611 (lane 4), Y9101 (lane 5) and CGS (lane 6) were separated with 15% polyacrylamide gel. The concentration was 1.5 x 106 spore equivalent. Arrowheads indicate homologous bands detected in all isolates.In order to investigate the diversity of N. bombycis, total protein, surface antigen, sequence of P30.4 (a putative surface protein) and sequence flanking region of 16S ribosomal RNA were compared using a variety of N. bombycis isolates and closely related microsporidia (Table 1). The results of this investigation are shown in Table 2. In protein analyses (SDS-PAGE and immunoblot assay), N. bombycis and other related microsporidia are readily distinguished from each other. In particular, four proteins were detected from all isolates of N. bombycis in Western analysis, suggesting that they were common antigenic determinants (Fig. 1). These bands were not detected in other microsporidia containing Nosema sp. NIS M11, Vairimorpha sp. NIS M12 and unidentified microsporidium TB-2M-H1 (data not shown). Partial sequences of 16S rRNA gene were highly conserved in N. bombycis group containing N. bombycis, N. furnacalis, N. richoplusiae and TB-2M-H1, and this group was related but separated from the Nosema apis group as reported previously (Fig. 2). In this way, species - specific characteristics were not detected in 16S rRNA sequence whereas the sequences of P30.4 gene were species-specific. The sequences were identical at amino acid level in N. bombycis, but differed from that of the unidentified microsporidium TB-2M-H1 (77% homology). Polyclonal antibodies against the spores of N. bombycis NIS 001 did not react with TB-2M-H1 (Hashiguchi, personal communication). This non-reactivity may be caused by the differences in amino acid sequences. It is known that N. bombycis produces two types of spores, which are called short coiled spores and long coiled spores. They are produced at different stages in the life cycle. Since P30.4 genes were transcripted throughout the infection stage (24 - 120 hrs postinoculation) in the process of host - parasite interaction, P30.4 may be a component of both spores. Localization and function of P30.4 in the spore formation process must be investigated in the future. Furthermore, it became clear that the ITS region between 16S and 5S rRNA genes were heterogeneous in an isolate. Gatehouse et al. also reported divergent sequences at the 3'-end of the large subunit (23S) rRNA gene in N. apis. The biological definition is unclear.
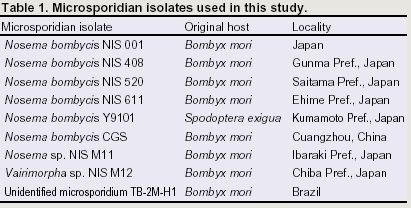
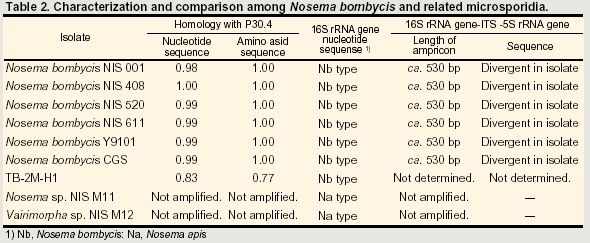
Fig. 2 Nosema / Vairimorpha phylogency based on small-subunit rRNA sequences.
Fig. 3 Comparison of genetic organization of rRNA genes among various organisms. In Vairimorpha necatrix and N. Apis, the RNA genes were arranged in the order 16S, ITS and 5.8S-23S. In theses organisms, the ITS region between 5.8S and 23S rRNA genes is not observed, and the 5' region of the 23S rRNA appears to correspond to the sequence of 5.8S rRNA. On the other hand, the organization of rRNA genes in N. bombycis was prokaryotic (16S rRNA gene, ITS and 5S) as shown in Fig. 3. Interestingly, it became clear that the organization of N. bombycis is quite different from that of Nosema apis. In fact, many researchers suggested that the genes Nosema contains the largest number of species and still represents a heterogeneous group, from which many species will have to remove to other genera. The result described above supported their opinions.
Finally, the diversity in the rDNA region in microsporidia might show the evolutionary route from prokaryotic type to eukaryotic type. The analysis of the rDNA region of N. bombycis, which is currently underway, will be necessary to clarify a number of the questions raised by the work reported here.