研究成果 Research Results
- TOP
- News
- Research Results
- Mysteries hidden in hepatic reprogramming
Mysteries hidden in hepatic reprogramming
Researchers get clearer picture of how just two proteins can make fibroblasts convert into liver-like cells 2020.10.28Research ResultsLife & Health
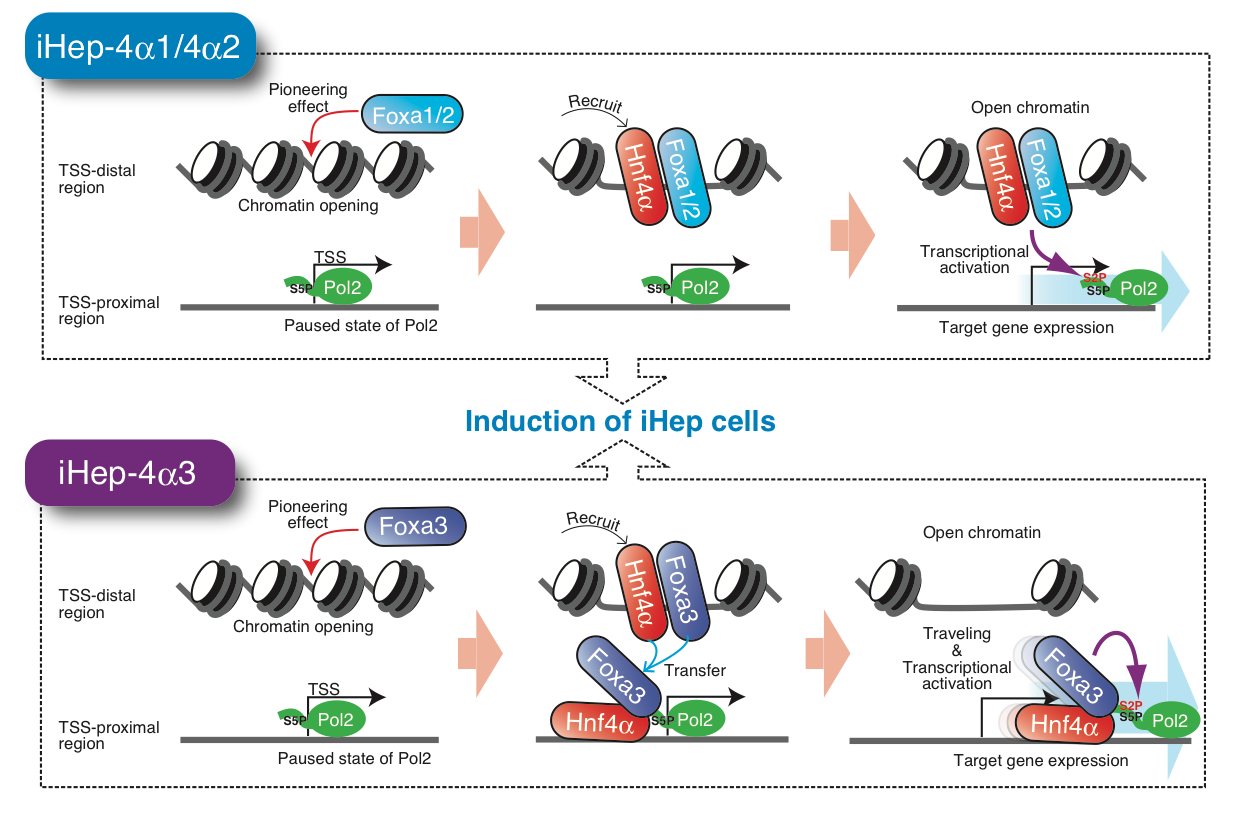
Foxa3 induces the expression of its target genes in a manner different from those of other family members, Foxa1 and Foxa2. iHep cells induced from fibroblasts using Hnf4α and Foxa1, Hnf4α and Foxa2, and Hnf4α and Foxa3 are designated iHep-4α1, iHep-4α2, and iHep-4α3, respectively. TSS: transcription start site, Pol2: RNA polymerase II.
Researchers in Japan have elucidated the molecular mechanisms of a process to turn cells called fibroblasts into those displaying functions similar to liver cells in mice, providing new insights for using such cells in medicine and for understanding liver diseases related to the disruption of cell formation.
Scientists have been exploring ways to generate various types of cells in the body for medical purposes, and one common approach is to induce cells to switch to an intermediate phase from which they can be programmed into any cell type.
However, Atsushi Suzuki, professor of Kyushu University’s Medical Institute of Bioregulation, previously succeeded in directly reprogramming mouse fibroblasts, a type of cells that form the structural network in tissues, into cells exhibiting properties of liver cells by introducing the right set of two transcription factors—proteins that play a key role in determining which genes of DNA are turned on and off.
Referred to as induced hepatocyte-like cells or iHep cells, the reprogrammed cells are promising for use in treating liver diseases and as a tool for understanding how liver cells form, but the detailed mechanisms for this conversion have remained a mystery.
“That just two transcription factors can make such sudden and drastic changes to a cell’s function is astounding,” says Suzuki. “After nine years of researching this mysterious phenomenon, we are finally getting a clearer picture of how this happens.”
Following an integrative analysis, Suzuki’s research team in collaboration with researchers at Kyoto University and the National Center for Global Health and Medicine have now unraveled the series of dynamic cellular state changes leading to this reprogramming.
Investigating three sets of two transcription factors—Hnf4α and one of Foxa1, Foxa2, or Foxa3—capable of triggering the conversion, the researchers found that all three Foxa proteins act as “pioneer factors” that open a specific region of DNA in tightly packed structures called chromatin and then recruit Hnf4α to further open the regions.
However, while Foxa1 and Foxa2 appear to stay in the same location, Foxa3 then immediately moves to the vicinity of transcription start sites—the beginning of DNA portions encoding target genes—with Hnf4α. There, Foxa3 binds to a group of molecules that reads the DNA to create RNA based on the DNA template and moves with this molecular machinery, known as RNA polymerase II, and Hnf4α along the DNA to efficiently induce transcriptional activation of target genes.
This unique feature of Foxa3 is indispensable for inducing conversion of fibroblasts to iHep cells using Hnf4α and Foxa3, and this knowledge could help to decipher the various roles of the Foxa proteins at different stages of development.
Understanding of the molecular mechanisms for iHep cell induction may provide insights into the medical applications of iHep cells and lead to elucidation of the molecular basis for regulating hepatocyte differentiation during liver development and regeneration and pathogenesis of liver diseases caused by disruption of hepatocyte differentiation.
###
For more information about this research, see “The dynamics of transcriptional activation by hepatic reprogramming factors,” Kenichi Horisawa, Miyako Udono, Kazuko Ueno, Yasuyuki Ohkawa, Masao Nagasaki, Sayaka Sekiya, and Atsushi Suzuki, Molecular Cell (2020). https://doi.org/10.1016/j.molcel.2020.07.012
Research-related inquiries
Atsushi Suzuki, Ph.D., Professor
Division of Organogenesis and Regeneration Medical Institute of Bioregulation, Kyushu University
Contact information can also be found in the full release.
- TOP
- News
- Research Results
- Mysteries hidden in hepatic reprogramming