研究成果 Research Results
- TOP
- News
- Research Results
- Barrier ahead? No problem for DNA replication
Barrier ahead? No problem for DNA replication
Study gives new insight into the mechanism underlying maintenance of genome stability in difficult-to-replicate regions 2020.12.22Research ResultsLife & Health
When copying sequences over six billion units long, one would expect the occasional obstacle or problem to arise. Yet somehow our cells manage to complete the job over and over again with perfect accuracy when replicating DNA by employing mechanisms to overcome these issues.
Now, researchers from Kyushu University have reported in the Journal of Cell Biology a new pathway for the DNA replication process to overcome such obstacles induced when DNA is tightly bound by proteins, giving new understanding about how our cells maintain genome stability in ‘difficult-to-replicate’ regions such as those found at the tips and linking points of chromosomes.
Known as DNA damage responses, mechanisms to continue the accurate replication of DNA despite conditions that hinder the process—broadly termed replication stress—have often been studied using external factors to inhibit DNA replication.
However, less is known about the response to tight interactions between chromosome DNA and specific binding proteins at internal ‘difficult-to-replicate’ regions such as centromeres and telomeres—the central linking points and tips of chromosomes, respectively.
By impeding the splitting of a DNA strand, such interactions can perturb progression of the fork formed as a DNA strand is split into two and the missing sides replicated to create two strands from one.
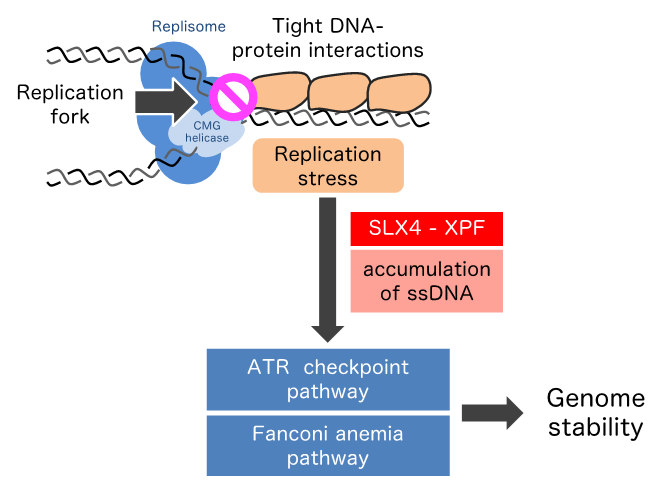
To elucidate the response to the replication stress evoked by tightly bound structures of DNA and protein, researchers led by Kazumasa Yoshida, researcher, and Masatoshi Fujita, professor, of Kyushu University’s Faculty of Pharmaceutical Sciences made site-specific replication fork barriers on the human chromosome by inserting areas called lacO arrays in the DNA and binding them using a bacterial protein known as LacI.
Comprehensively investigating how the DNA damage response is activated when replication is blocked by the barriers, the researchers found a novel signaling pathway in which a scaffold protein SLX4 and a structure-specific endonuclease XPF mediate recruitment of ATR, FANCD2, and RAD52 to the stalled fork.
Furthermore, they demonstrated that the SLX4-XPF-ATR axis contributes to complete replication of the lacO array.
“This study provides a comprehensive picture of a process by which human cells manage nucleoprotein obstacles ahead of replication forks, shedding light on mechanisms underlying the DNA replication and repair, as well as the genomic instability associated with oncogene-induced replication stress,” says Yoshida.
###
For more information about this research, see “SLX4–XPF mediates DNA damage responses to replication stress induced by DNA–protein interactions,” Riko Ishimoto, Yota Tsuzuki, Tomoki Matsumura, Seiichiro Kurashige, Kouki Enokitani, Koki Narimatsu, Mitsunori Higa, Nozomi Sugimoto, Kazumasa Yoshida, and Masatoshi Fujita, Journal of Cell Biology (2020). https://doi.org/10.1083/jcb.202003148
This research was supported in part by grants from the Japan Society for the Promotion of Science (JP15K18478), the Fukuoka Foundation for Sound Health Cancer Research Fund, and the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
This release is also available in Japanese.
Research-related inquiries
Masatoshi Fujita, Professor
Kazumasa Yoshida, Researcher
Faculty of Pharmaceutical Sciences
Contact information can also be found in the full release.
- TOP
- News
- Research Results
- Barrier ahead? No problem for DNA replication