研究成果 Research Results
- TOP
- News
- Research Results
- A new way to map how cells choose their fate
A new way to map how cells choose their fate
Researchers present an innovative computational method based on modern mathematics to shed light on cell state dynamics and developmentAssociate Professor Kazumitsu Maehara
Faculty of Medical Sciences
2025.12.29Research ResultsLife & HealthMath & Data
Fukuoka, Japan—Researchers from Kyushu University have developed an innovative computational method, called ddHodge, that can reconstruct the complex dynamics of how cells decide their fate. As reported in Nature Communications, this approach paves the way for a deeper understanding of the biological processes involved in development, regeneration, and disease.
Understanding how a developing cell chooses its destiny, such as differentiating into a nerve cell or a muscle cell, is a central challenge in biology and medicine. To study these mechanisms, scientists often rely on single-cell RNA sequencing (scRNA-seq)—a technology that reveals which genes are active within individual cells. While powerful, scRNA-seq is destructive, meaning that it can only provide one-time snapshots of cells, but not the evolution of their states over time.
Computational methods like RNA velocity have begun to tackle this limitation by inferring both the immediate future direction of a cell and the “speed” at which it advances toward it. However, a cell’s state is defined by innumerable genes, placing it in a complex, high-dimensional space. As current techniques cannot accurately represent this full space, they compress it into far fewer dimensions, inevitably losing important information about the data geometry. As a result, it is impossible to consistently assess the stability of a cell state—that is, one cannot distinguish a highly plastic, unstable cell at a branching point from one that is deeply committed and stable.
Against this backdrop, Associate Professor Kazumitsu Maehara from Kyushu University’s Faculty of Medical Sciences and Professor Yasuyuki Ohkawa from Kyushu University’s Medical Institute of Bioregulation have developed ddHodge, a geometry-preserving method that can more accurately reconstruct cell state dynamics.
“My background is in statistical science, and during my graduate training, I was exposed to HodgeRank, a method used in ranking problems such as PageRank,” says Maehara. “When I later moved into life-science research, I realized that the same mathematical idea could help interpret the complex, high-dimensional transitions in single-cell data.”
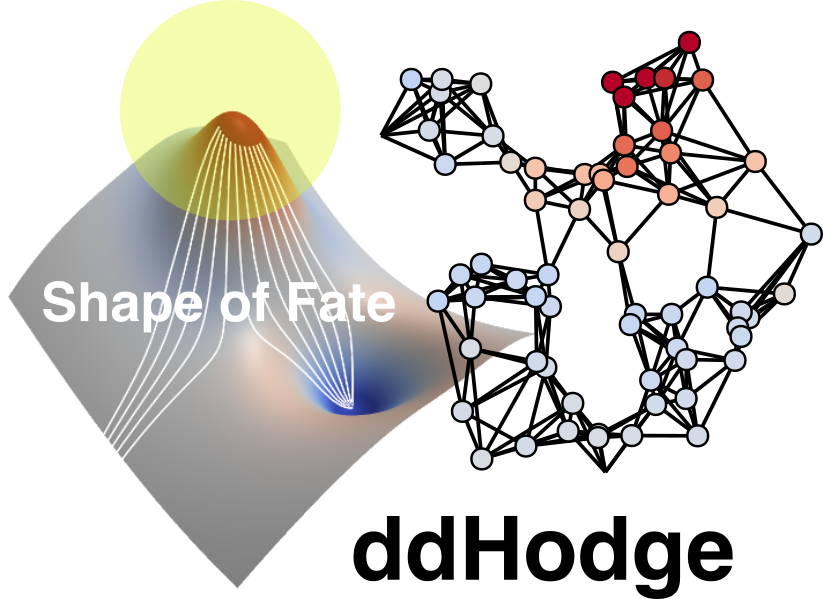
The left panel depicts a potential-like landscape, where white streamlines represent the inferred trajectory of cell states across time. Although single-cell RNA sequencing captures only static snapshots of cells, ddHodge reconstructs the patch-worked dynamical structure by connecting many local “nows” into a coherent global picture. The right panel shows a graph-based representation of high-dimensional cell states, on which local geometry is preserved to quantify key dynamical properties such as stability, divergence, and rotational components. https://github.com/kazumits/ddHodge.jl
Their technique is based on Hodge decomposition, a powerful mathematical theorem, which they used to break down cells’ motion across a landscape of possible states into three fundamental and measurable components. The first is the gradient, which is the overall directional flow across the landscape. The residual contains the curl and the harmonic components, which account for cyclical or rotational flows and can thus reveal repeating processes like the cell cycle.
“ddHodge can be viewed as an effort to adapt techniques and concepts developed in modern mathematical sciences, such as differential geometry and numerical computation, to the practical demands of life science data analysis,” explains Maehara. The proposed framework utilizes geometric principles to approximate how cell states “move” on a lower-dimensional structure while preserving the shape information embedded in high-dimensional data, which is normally lost in standard methods that rely on dimensionality reduction.
When applying ddHodge to scRNA-seq data from approximately 46,000 mouse embryonic cells, the researchers found that over 88% of the gene expression dynamics during early embryonic development could be explained by the gradient component. This substantiated, with real-world data, the long-standing concept in developmental biology that cells differentiate by moving toward stable states and diverging away from “branching points.” Moreover, by focusing on these unstable points, the researchers were able to identify key genes that drive or maintain cell state stability as cells commit to a lineage.
The researchers also evaluated ddHodge’s performance using data simulations, revealing that even when given partial or noisy data, ddHodge was able to reliably reconstruct cell state dynamics, with around 100 times more accuracy than other conventional approaches.
Overall, ddHodge provides a reliable way to identify critical biological moments, such as the exact timing and location of cell fate decisions. “ddHodge can quantitatively describe, within a high-dimensional space, in which direction, how fast, and how stably cells change. We expect it to contribute broadly to understanding diverse biological phenomena, including embryonic development, tissue regeneration, and cancer progression,” adds Maehara. This tool could support the early detection of cell states relevant to disease states or regeneration, as well as help scientists analyze large-scale datasets used in pharma and biotech discovery pipelines.
Notably, ddHodge has many potential applications beyond biology and medicine. The researchers believe it could be used to provide insights into other complex processes that change over time, including material degradation, climate patterns, and socioeconomic behavior. Thus, ddHodge exemplifies how concepts from modern mathematics can be used to gain insights into processes and systems that would otherwise be obscured in giant high-dimensional datasets.
###
For more information about this research, see “Geometry-preserving vector field reconstruction of high-dimensional cell-state dynamics using ddHodge,” Kazumitsu Maehara and Yasuyuki Ohkawa, Nature Communications, https://doi.org/10.1038/s41467-025-67782-6.
Research-related inquiries
Yasuyuki Ohkawa, Professor
Medical Institute of Bioregulation
Contact information can also be found in the full release.
- TOP
- News
- Research Results
- A new way to map how cells choose their fate